Part 1: Ion-Exchange Chromatography
Introduction
Therapeutic synthetic oligonucleotides represent a significant advancement in the field of molecular medicine, offering new avenues for the treatment of a wide range of diseases that were previously challenging to manage or ‘undrugabble’. Emerging in the late 20th century, this innovative class of therapeutics includes small, single-stranded DNA or RNA molecules designed to modulate gene expression or affect protein function.
Therapeutic agents such as antisense (ASOs) and splice switching oligonucleotides, small interfering RNA (siRNA), aptamers, antagomirs and microRNA (miRNA) mimicry or inhibition allow for precise targeting of specific genetic sequences. This specificity not only enhances the efficacy of these therapies but also minimizes off-target effects, a common challenge in traditional drug development.
The development of therapeutic oligonucleotides from concept to clinical application has faced numerous challenges, including delivery mechanisms, stability, and immune system recognition. However, through chemical modification, nanoparticle encapsulation and other innovations, significant strides have been made. To date, eighteen oligonucleotide-based drugs have received FDA approval for treating conditions such as spinal muscular atrophy, Duchenne muscular dystrophy, and familial hypercholesterolemia, marking the emergence of a new era in precision medicine. Continued research highlights the potential of therapeutic oligonucleotides not only for rare genetic disorders but also for common diseases, including cancers and viral infections, the latter of which has been accelerated following the successful development of mRNA vaccines to combat the SARS-CoV-2 pandemic.
Oligonucleotide Analysis – Chromatographic Techniques
Oligonucleotide therapeutics defy classification as either small molecule or biologic entities as they embody traits of both groups, sharing the complex chemistry typical of biologics, yet are commonly synthesised like small molecule therapies even though their molecular weight can vary widely. This being the case, critical quality attributes associated with identity, potency, quality, and purity still need to be assessed and, due to the oligonucleotide chemical complexity, several analytical techniques are employed to achieve these measurements. Table 1 contains a summary of a selection of the chromatographic approaches to oligonucleotide critical quality attribute assessment and a small number of applications and potential problems;
| Technique | Applications | Comments |
| Size Exclusion Chromatography |
|
|
| Reversed Phase HPLC(MS) |
|
|
| Ion Pair HPLC(MS) |
|
|
| Strong Anion-Exchange (SAX) HPLC |
|
|
| HILIC |
|
|
Table 1: Characterization of impurities generated during solid phase oligonucleotide synthesis (SPOS) is a crucial part of pharmaceutical process development and quality control.
Oligonucleotide Synthesis
A typical synthetic production cycle of phosphoramidite oligionucleotides (just one of several possible methods of production) is shown in figure 1;


Figure 1: 5 step synthesis of oligonucleotides by the H-Phosphonate Method
Synthesis is based on stepwise addition of nucleotide residues to the 5'-terminus of the growing chain until the desired sequence is assembled. To mitigate the occurrence of non-targeted reactions, protective groups are employed to modify nucleosides, exemplified by the conjugation of a dimethoxytrityl (DMT) moiety to the 5’-hydroxyl segment of the sugar component as shown in Figure 1, which can be removed (deprotected) prior to the next coupling cycle. The process is susceptible to the accumulation of errors, culminating in the generation of contaminants and degradation by-products such as the formation of structurally analogous impurities, notably, longer oligomers (longmers, n+x) and shorter oligomers (shortmers, n–x), which are a number of bases more or less than the desired full-length product.
Therapeutic oligonucleotides frequently undergo modifications to increase their stability or to enhance pharmacokinetics. These modifications can be applied to the backbone, base, sugar moieties, or through structural conjugation. Typically, oligonucleotides possess phosphodiester linkages (PO), however a common modification involves replacing the oxygen atom with sulphur to create a phosphorothioate linkage (PS), which enhances stability against nucleases and reduces nucleolytic degradation (figure 2). PS oligonucleotides are widely utilized in both in vivo and in vitro applications due to their increased resilience compared to PO oligonucleotides. The nucleophilic hydroxyl group at the 2'-ribose position is frequently modified with an O-methyl (2'-OMe) group, which also boosts resistance to nuclease digestion (Figure 2).


Figure 2: Oligonucleotide modifications. (Courtesy of YMC Europe GmbH, Dinslaken, Germany)
Strong Anion Exchange Chromatography
Due to the negatively charged phosphate groups in the backbone of oligonucleotides Ion Exchange (IEX) chromatography is commonly used for detecting structural variations. At pH 7-8 the oligonucleotide will have negative charges proportional to the number of nucleotides minus 1, and in this first of a series of Technical Notes on oligonucleotide analysis, we highlight some important applications using the ion exchange technique, which uses electrostatic interaction between the analyte and stationary phase to affect retention.
In anion exchange separations of oligonucleotides, salt gradients are used to achieve selective elution of the oligonucleotide from the stationary phase, by disruption of the analyte stationary phase interaction with increasing concentration of buffer counter ion as shown in figure 3.


Figure 3: Elution mechanism for oligonucleotides in anion-exchange chromatography (Courtesy of YMC Europe GmbH, Dinslaken, Germany)
The pH and counter ion choices used in anion exchange chromatography of oligonucleotides is dependent upon the nature of the species being analysed. Often, species with lower net negative charge, such as the phosphodiester oligonucleotides can be chromatographed at near to neutral pH with simple salts such as NaCl. For more highly negatively charged species, such as the phosphorothioate oligonucleotides or for longer oligonucleotide species, the higher negative charge per analyte would lead to an unmanageable increase in retention and reduction in efficiency under the conditions mentioned previously. For these species, analysis at higher pH, typically pH 12, and the use of more aggressive counter ions such as NaClO4 are necessary to affect elution with good peak shape. Ion-exchange analysis of oligonucleotides can also be carried out at high pH to eliminate unwanted secondary interactions such as Watson-Crick hydrogen bonding, significantly sharpening the chromatographic peak, and to enable resolution of problem sequences such as self-complimentary sequences and poly-G stretches. When working at elevated pH, it is necessary to use strong anion-exchange functionalised packings to ensure the surface on the stationary is fully charged at pH12-13.
Figure 4 shows an optimised ion exchange separation of single stranded DNA, RNA and 2’OMe all with single base differences.


| Column: | YMC BioPro IEX QF (5 µm) 100 x 4.6 mm ID |
| Eluent: | A) 10 mM NaOH, B) 10 mM NaOH containing 1.0 M NaClO4 |
| Gradient: | 25–55%B (0–15 min), 100%B (15–20 min) |
| Flow rate: | 1.0 mL/min |
| Temperature: | 25 °C |
| Detection: | UV at 260 nm |
| Injection: | 4 µL (5 nmol/mL each) |
| Samples: |
DNA 20 and 21 mer (samples 1 and 2), RNA 20 and 21 mer (samples 5 and 6), 2'-OMe RNA 20 and 21 mer (samples 3 and 4) |
Figure 4: Separation of n from n-1-mer DNA, RNA and 2’-OMe functionalised oligonucleotides (Courtesy of YMC Europe GmbH, Dinslaken, Germany)
The separation of figure 4 clearly demonstrate the utility of the ion exchange technique for the analysis of oligonucleotide sequences which differ by only one nucleoside, including RNA, DNA and 2’-O-methyl species. As the optimised eluent system has a starting pH of around 12, the BioPro IEX phase from YMC, which is a non-porous polymethacrylate support material with an amide functionalised strong anion exchange bonded phase, is excellent at resisting chemical degradation. The use of sodium hydroxide, rather than more traditional Tris buffer, alongside the use of the perchlorate counter ion rather than a chloride, significantly improves peak shape. Changing the buffer and counter ion type are important variables in method development for oligonucleotide analysis. While higher-pH mobile phase buffers may improve peak shapes of shorter oligonucleotides, longer oligonucleotides can suffer from pH-mediated degradation of the product, and should be monitored to determine the maximum temperature and pH combination to use before purification related degradation is induced.
Chromatographic Optimisation
The analysis of oligonucleotides is challenging chromatographically, however, the chromatographer has several tools within their armoury to enable better efficiency and sensitivity, as shown in figure 5.




| Column: | YMC BioPro IEX QF (5 µm) 100 x 4.6 mm ID |
| Eluent (Top): | A) 10 mM NaOH B) 10 mM NaOH containing 1.0M NaClO4 |
| Eluent (Bottom): | A) 10 mM NaOH/methanol (70/30) B) 10 mM NaOH containing 1.0M NaClO4/methanol (70/30) |
| Gradient (Top): | 32–80%B in 24 min |
| Gradient (Bottom): | 40–100%B in 6.3 min |
| Flow rate: | 1.0 mL/min |
| Temperature: | 25°C (top) / 60°C (bottom) |
| Detection: | UV at 260 nm |
| Injection: | 2 µL (10 nmol/mL) |
| Samples: | RNA 20 and 21 mer All PS (samples 13 and 14) |
Figure 5: Optimisation of phosphorothioate oligonucleotides (20-mer and 21-mer) separation (Courtesy of YMC Europe GmbH, Dinslaken, Germany)
The much-improved separation efficiency, resolution and sensitivity in the lower chromatogram of figure 5 is obtained via addition of an organic modifier, increasing analysis temperature and a steeper gradient. The addition of the organic modifier acts to suppress the formation of secondary structures which causes unnecessary peak broadening. The optimisation process to achieve the lower chromatogram was not achieved in a single iteration, however the example serves to demonstrate some of the key variables available in the ion exchange separation of closely related oligonucleotides.
Separating Larger Oligonucleotides
With smaller oligonucleotides (n=10-30), separation of n from n-1 species is relatively straightforward, however as molecular weight increase (n>50), the relative charge density between n and n-1 species becomes much smaller and the kinetics of mass transfer of analyte into and out of the stationary phase tends to lead to appreciable peak broadening. Strategies to solve these issues include working at elevated temperatures, the use of organic modifiers and the optimisation of the pore size to ensure the most efficient mass transfer of larger oligonucleotides.
Figure 6 from Agilent Technologies demonstrates the separation of poly-T-oligonucleotides at slightly lower pH but using a sacrificial base reagent (TEAA) to overcome some of the peak shape issues due to secondary interactions, that otherwise may be present at close to neutral pH values. The application uses a Agilent PL-SAX polymeric column with particle and pore size optimised for the separation of a wide range of oligonucleotides molecular weight (length). The figure shows the high resolution of oligonucleotides to a 50-mer as well as a purification of a 91-mer, both using the same stationary phase but with a longer column used to achieve the required resolution of a 91-mer species.
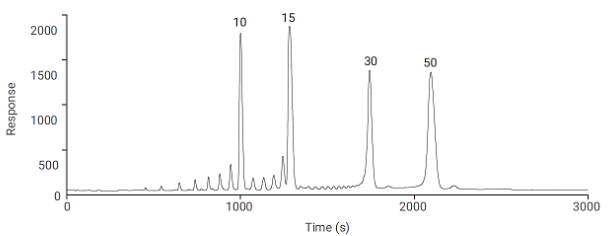



| Top: |
Separation poly-T-oligonucleotide size standard spiked with a 10mer, 15mer, 30mer, and 50mer |
| Column: | Agilent PL-SAX 1000Å, 8 μm, 50 × 4.6 mm |
| Eluent A: | 7/93 v/v acetonitrile/0.1 M TEAA, pH 8.5 |
| Eluent B: | 7/93 v/v acetonitrile/0.1 M TEAA, 1 M ammonium chloride pH 8.5 |
| Gradient: | 0 to 40% B in 10 min, 40 to 70% B in 14 min, 70 to 100% B in 25 min |
| Flow rate: | 1.5 mL/min |
| Temperature: | 60 °C |
| Bottom: |
Purification of a large, 91mer, synthetic oligonucleotide |
| Column: | Agilent PL-SAX 1000Å, 8 μm, 150 × 4.6 mm |
| Eluent A: | 7/93 v/v acetonitrile/0.1 M TEAA, pH 7.0 |
| Eluent B: | 7/93 v/v acetonitrile/0.1 M TEAA, 3.24 M ammonium acetate pH 7.0 |
| Gradient: | 0% B for 2 min, 0 to 100% B in 20 min |
| Flow rate: | 1.5 mL/min |
| Temperature: | 60 °C |
Figure 6: High resolution separation of a poly-T-oligonucleotide size standard spiked with a 10mer, 15mer, 30mer and 50mer (top) and purification of a large, 91mer, synthetic oligonucleotide(bottom) (Courtesy of Agilent Technologies, Santa Clara, California, USA)
Note that the top separation in Figure 6 is obtained using a multi-step gradient. When separating wide ranges of oligonucleotide lengths, it is useful to alter the rate of change of the salt gradient in order to improve resolution of the longer species, typically the gradient slope is lowered as molecular weight increases.
Figure 7 shows the separation of oligodeoxythymidine samples with 12–60 bases including several sequence failures, incompletely deprotected and otherwise modified species using a polymeric DNAPac PA200 RS strong anion exchange column. This column has a pellicular strong anion exchange coating applied to a non-porous 4mm polymeric particles which produces high efficiency separations.


| Column: | DNAPac PA200 RS (4 μm), 4.6 × 150 mm |
| Mobile Phase: | A) 40 mM Tris, pH 8 / B) 40 mM Tris, pH 8, 1.0 M NaCl |
| Gradient: | 41 to 65% B in 8.4 minutes (curved gradient) |
| Flow rate: | 1.0 mL/min |
| Temperature: | 30 °C |
| Diode-array | 260 nm detector |
Figure 7: Separation of deoxythymidine oligodeoxynucleotides revealing 49 full-length synthetic oligonucleotides (ONs), with eight failure sequences and 26 modified or incompletely deprotected oligonucleotides (Courtesy of Thermo Fisher Scientific Inc., Waltham, Massachusetts, United States)
It is interesting to note that the high-resolution separation of figure 7 is achieved using a curved (concave) gradient in which the rate of change of ionic strength is continually lowered over time, rather than by employing a multi-step gradient. Many modern instruments are capable of programming non-liner gradients and this can be very useful in the analysis of higher molecular weight oligonucleotides.
Analysis of Structural Variants
Whilst it is impossible to show the full range of analyses of oligonucleotide impurities and sequence failures using Element Laboratory Solutions products, it is worthwhile considering an example of impurities based on structural variants of homo-nucleotides, that is nucleotides of the same sequence length but differing chemistry or structure. In this example, aberrant 2’,5’-linkages were introduced at specified positions along a mixed-base 21-mer comprising the antisense strand of an eGFP siRNA oligonucleotide pair. These aberrant linkages may be introduced at positions in siRNA sequences where an unprotected 2’ hydroxyl is available for phosphoryl-migration and oligonucleotides with aberrant linkages at different positions were mixed at different concentrations (to aid identification) and chromatographed as shown in figure 8.


| Column: | DNAPac PA200 (8 μm) 4.0 × 250 mm |
| Mobile Phase: | A) 40 mM Tris, pH 8 / B) 40 mM Tris, pH 8, 1.0 M NaCl |
| Gradient: | 49 to 71% B in 19.5 minutes |
| Flow rate: | 1.0 mL/min |
| Temperature: | 30 °C |
| Diode-array | 260 nm detector |
Figure 8: Resolution of a 21-base eGFP antisense strand with all normal 3’,5’ linkages and five different ONs of the same sequence, but with 2’,5’-linkages inserted at positions 1, 5, 10, 15, and 20 (Courtesy of Thermo Fisher Scientific Inc., Waltham, Massachusetts, United States)
Figure 8 shows the positions of the aberrant linkages with the 0 marker indicating the species all (normal) 3’-5’ linkages. All species, which have the same sequence and length, are separated within 10 minutes using a long column and a shallow linear gradient which is optimised for resolution of all analytes. The mechanism for this recognition is not fully defined, however it is considered that the introduction of aberrant linkages alters the solution confirmation of each oligonucleotide. The ability of anion exchange chromatography to differentiate between such closely related analytes, highlights the power of anion exchange chromatography in the analysis of oligonucleotide impurities.
Key factors in Purification of Oligonucleotides by HPLC
When purification of various oligonucleotide species is the analytical goal, there are several further variables which need to be considered. Typically, in purification, one wishes to recover as much material as possible per injection and as such, the loadability of the stationary phase must be considered. At analytical scale, non-porous particles represent the best option and yield high efficiency and sharp peaks at low analyte loading. However, at the preparative scale, porous materials are often a better option as the higher surface area they offer, improves loadability. Whilst loadability of the porous materials is higher, efficiency can be reduced compared to the non-porous material due to the mass transfer processes associated with analyte diffusion into and out of the porous structure. This principle is demonstrated with YMC BioPro IEX resins in figure 9.


| Column: | YMC BioPro IEX SF/SP (5 μm) 50 x 4.6 mm ID |
| Eluent: | A) 20 mM NaH2PO4-Na2HPO4 (pH 6.8) / B) 20 mM NaH2PO4-Na2HPO4 (pH 6.8) containing 0.5 M NaCl |
| Gradient: | 0–100%B (0–60 min) |
| Flow rate: | 0.5 mL/min |
| Temperature: | 25 °C |
| Detection: | UV at 280 nm |
| Injection: | 100 μL |
| Sample: | Lysozyme (180mg on column) |
Figure 9: Loadability of YMC BioPro IEX porous and non-porous ion-exchange media (Courtesy of YMC Europe GmbH, Dinslaken, Germany)
The fronting appearance of the lysozyme peak in figure 9 indicates that the non-porous stationary phase media is overloaded and the purified fraction would be collected over a wider timeframe, ultimately resulting in more dilute fraction with a higher solvent volume to remove in order to recover the purified material. The sharper peak obtained with the porous media ultimately results in a higher concentration of the recovered material within the collected fraction.
A further consideration when selecting preparative media is particle size as column backpressure increases with decreasing particle diameter. Whilst the efficiency of the separation (and ultimately therefore the resolution) decreases with increasing particle size, ultimately recovery (purity) of each fraction needs to be balanced with loadability and the back pressure restraints of the equipment being used. YMC BioPro IEX SmartSep resins are available in a range of particle sizes from 10 to 30mm to enable scale up and scale down depending upon recovery requirements, however, crucially the retention profile of oligonucleotides on this series of resins shows excellent fidelity, significantly easing the scaling process. The relative efficiency of these resins and the retention fidelity is shown in figure 10.


| Column: | YMC BioPro IEX SmartSep 100 × 4.6 mm ID |
| Eluent: | A) 10 mM NaOH / B) 10 mM NaOH + 1 M NaClO4 |
| Gradient: | 25–40%B (0–30 min), 40–100%B (30–35 min) |
| Flow rate: | 1 mL/min |
| Temperature: | 25 °C |
| Detection: | UV at 260 nm |
| Injection: | 5 μL |
| Sample: | Trityl-off DNA 20mer |
Figure 10: Loadability and retention time reproducibility of 10 and 30mm particle diameter YMC BioPro IEX SmartSep resins (Courtesy of YMC Europe GmbH, Dinslaken, Germany)
Oligonucleotide Analysis Workflow solutions
Element Laboratory Solutions also offer many workflow products to support the ion-exchange analysis of oligonucleotides, including the innovative TOP DNA and TOP RNA purification products. These trityl-on oligonucleotide purification (TOP) products use a 6-step extraction process to achieve greater than 90% yield and purity for nucleotides of up to 140-mer sequences. Figure 11 shows the purification of a 70-mer DNA oligonucleotide with 96% purity and 93% yield.


| Column: | PLRP-S 5 μm, 4.6 x 150 mm |
| Mobile Phase: | A) 100 mM TEAA, pH 7 / B) Acetonitrile |
| Gradient: | A/B (95:20) from 0 to 20 minutes |
| Flow rate: | 1.0 mL/min |
| Temperature: | Ambient |
| Detection: | UV @ 256 nm |
Figure 11: Purification of a 70mer DNA sequence using 150 mg TOP-DNA tubes (Courtesy of Agilent Technologies, Santa Clara, California, USA)
Element Lab Solutions also offer oligonucleotide resolution standards containing 14, 17, 20 and 21-mer synthetic oligonucleotides for assessing column or separation performance. DNA ladder standards are also available for testing column selectivity and reproducibility and both standards are available as lyophilized solids and come with a certificate of analysis.
When using high pH eluents for ion exchange separations, any change in the mobile phase pH due to ingress of carbon dioxide can lead to significant changes in selectivity and resolution of the oligonucleotides. To prevent ingress of carbon dioxide, we offer Safety Caps from SCAT which hermetically seal the eluent bottle and prevent changes in chromatography.
Summary
In this first technical note of our series on Oligonucleotide Analysis, we have demonstrated the power of Ion Exchange Chromatography in the analysis of longmers, shortmers, structural impurities and conformers. Under the correct analysis conditions, high efficiency and high sensitivity separation of oligonucleotide species is possible, however the choice of eluent pH, salt species, salt gradient, temperature and the use of an organic modifier all play an important role is obtaining a fit for purpose separation. When purification of the oligonucleotide is required, particle size and morphology also need to be considered.
Element Laboratory Solutions offer a broad selection of Strong Anion Exchange columns from world-class manufacturers, alongside workflow products to ensure successful IEX separations to drive quality control, production, or development decisions in your laboratory.
Contact us to discuss our range of products and the ways in which we can help to develop and optimise your oligonucleotide analysis.