Part 2: Reversed Phase Chromatography
Introduction
Polymeric sequences of nucleotides (DNA, RNA, and their analogs) called oligonucleotides are used in a wide range of applications, such as PCR primers, silencing RNA experiments, diagnostic probes, and innovative medicinal treatments. AgBio businesses are also investigating oligonucleotides as novel types of tailored crop protection. Companies that invest in research and development, as well as manufacturing, need reliable analytical tools and solutions to meet regulatory requirements and expedite development.
The phrase "therapeutic oligonucleotides" describes a wide range of nucleic acid structures, including gapmers, aptamers, antisense oligonucleotides (ASOs), mRNA mimics, antagomirs, and agomirs, and splice-switching oligonucleotides (SSOs). Based on their size and complexity, therapeutic oligonucleotides are typically divided into three groups: short single-stranded oligonucleotides (siRNA), large single-stranded oligonucleotides (aptamers), and short double-stranded oligonucleotides (ASOs, SSOs, gapmers, antagomirs, and agomirs). Longer oligonucleotides are produced via the preferred enzymatic in-vitro transcription (IVT) method, while solid-phase synthesis can produce short oligonucleotides.
In order to achieve GMP regulatory standards for oligonucleotide identity and sequence confirmation as well as for finding and characterizing contaminants associated to synthesis, LC-MS based analytical procedures are essential. Ion-Pairing Reversed Phase (IP-RP) liquid chromatography is an important analytical technique since it offers excellent selectivity, high resolution, and MS compatibility. MS analysis is now more important than ever to verify identity and sequence because of the expanding variety of modified nucleotides, dye/quencher labels, and conjugations (GalNac, cholesterol, mAbs, etc.).
Oligonucleotide Analysis – Chromatographic Techniques and Synthesis
An iterative four-step synthesis procedure is used to create synthetic oligonucleotides, adding nucleotides one after the other until the desired output is produced. Despite the fact that this procedure is well-understood and effectively executed, chromatographic analysis is used extensively for impurity profiling of longmers, shortmers and structural variants. The hydrophobicity of the bases and the negatively charged phosphate backbone of the oligo structure allows for five different LC modes to be employed for investigation. While the nucleobases provide a degree of hydrophobicity, it is probably more typical to use the reversed phase mode to separate or purify the dimethoxytrityl (DMT)-on (protected) from the DMT-off (deprotected) species. Although anion exchange chromatography is well suited to the separation of oligonucleotide charge variants, it is not well suited for MS.
For a full list of Chromatographic Techniques and a schematic of the synthetic production cycle, please refer to Part 1 of this series.
Ion-Pair Reverse Phase (IP-RP) Chromatography
Ion-Pair Reverse Phase Chromatography involves two fundamental biophysical characteristics of DNA: hydrophobicity resulting from the heterocyclic bases and polarity caused by the negatively charged phosphate backbone. The oligonucleotide chain is retained via the hydrophobic interaction between the stationary phase sorbent and the DNA bases, while the charge on the DNA is neutralized using a conventional ion-pairing agent (such as triethyl amine, or triethylamine acetate (TEAA), which contains a positively charged tertiary amine). Nonetheless, a significant problem with TEAA is its substantial sequence-specific retention bias, which becomes more pronounced as the oligonucleotide sequence becomes more complicated. Therefore, TEAA may be effective for homo-oligomers but not for hetero-oligomers. The proven technique of ion pair reversed phase chromatography serves as the foundation for the separation of detritylated (DMT-off, deprotected) synthetic oligo materials.
Selecting an Appropriate Stationary Phase:
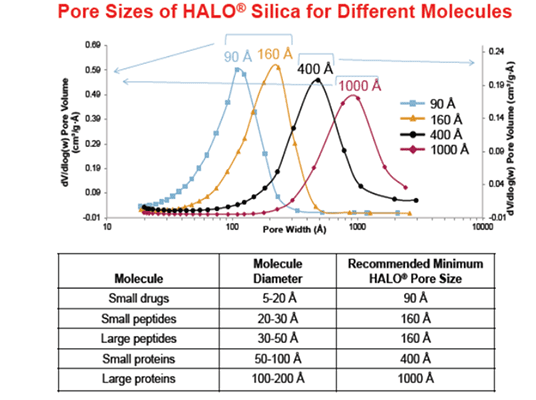
Specialized columns are needed for these separations since it is challenging to retain and separate highly polar molecules, such as short oligonucleotides, on standard reversed phase columns. The desired column characteristics may include larger pore diameter, temperature and pH stability and resistance to phase collapse/de-wetting in 100% aqueous mobile phases. Due to their resilience at high temperatures and wide usable pH range, hybrid particle columns offer method development flexibility, while silica-based polar columns show improved retention for more polar analyte molecules. Since pore size affects oligonucleotide diffusivity and, consequently, peak shape, it should also be considered as a significant parameter.


| Column: | 50 x 2.1 mm ID |
| Eluent: | A) 100 mM or 10 mM Triethylamine-acetic Acid (pH 6) B) A/Acetonitrile (80/20) |
| Gradient: | 50 – 65% B (0 – 20 min) |
| Flow rate: | 0.21 mL/min |
| Temperature: | 35 °C |
| Detection: | UV at 260 nm |
| Injection: | 2 µL (5 nmol/mL) |
| Sample: | Oligodeoxythymidylic acid [d(pT)2-20] |
Figure 1: The separation of oligo(deoxythymidylic acids), d(pT)2-20’ which was compared using triethylammonium acetate (TEAA) buffer, under the same gradient conditions, on different stationary phases (Courtesy of YMC Europe GmbH, Dinslaken, Germany)
The chromatograms above highlight the importance of selecting the correct concentration of buffer. 100 mM buffer shows significant improvement over 10 mM buffer, in both retention and resolution of all oligonucleotide sequences. This is true for each of the column stationary phases tested. These improvements can be attributed to more efficient ion-pairing between the charged oligonucleotide and the ion-pairing agent, when the ion-pairing agent is present at higher concentrations.


| Column: | HALO ES-C18, 2.1 x 150 mm, 1000Å, 2.7 µm |
| Eluent: | (A) 100mM TEAA Buffer pH 7.0, (B) 80% A, 20% AcN |
| Gradient: | 40 – 62.5%B (0-20 min), 62.5 – 100%B (20-22 min), 100%B (22-23 min) |
| Flow rate: | 0.2 mL/min |
| Temperature: | 60 °C |
| Detection: | UV |
| Sample: | Oligonucleotide Ladder, 20 to 100 bases |
Figure 2: The effect of Pore Size comparison on small and large oligonucleotides using a 20/100 oligonucleotide ladder (Courtesy of Advanced Materials Technology Inc (Halo) , Wilmington, DE)
| Manufacturer | Description | Phase | Particle Type | Particle Size | Pore Size | pH Range | Max. Temperature |
| Agilent | Advance Bio | RP | Superficially Porous | 2.7, 4 µm | 120 Å | 3 - 11 | 65°C |
| Avantor | Ace Excel Oligo | RP | Silica | 1.7, 3 µm | 90 Å | 1.5 – 11.5 | 100°C |
| Thermo | DNAPac 200/RS | IEX | Polymeric | 4 µm | Non-porous | 2.5 – 12.5 | 85°C |
| Thermo | DNAPac RP | IEX | DVB | 5 µm | Proprietary Wide Pore | 0 - 14 | 110°C |
| YMC | Accura Triart | RP | Hybrid | 1.9, 3, 5 µm | 120 Å | 1 - 12 | 90°C |
| YMC | BioPro | IEX | Resin | 3.5, 5 µm | 1000 Å , non-porous | 2 - 12 | 80°C |
Table 1: The range of Oligonucleotide columns available from Element Laboratory Solutions (Courtesy of Element Laboratory Solutions, Strathaven, UK)
Selecting an appropriate ion-pairing mobile phase
Whichever mobile phase system is chosen, the use of LC-MS grade solvents and additives is recommended when available, specifically when MS detection is required. Due to the negatively charged phosphate backbone, MS is typically run in negative mode, which results in the presence of sodium and potassium adducts. Those familiar with MS detection of oligonucleotides will recognize this as a significant problem. Although it should be noted that different vendors assay their reagents differently, it’s clear that lower purity additive is much more susceptible to adduct formation (for example, 99% pure HFIP versus 99.8% pure HFIP), which results in reduced sensitivity of each ion in MS detection.
It is wise to run a standard to benchmark system and column performance prior to analysis of samples. Make sure your standard performs always as expected, especially if your instrument has been idle for a while or if you're working with samples that you haven’t previously analysed.


| Column: | AdvanceBio Oligonucleotide, 2.1 x 50 mm (p/n 659750-702) |
| Mobile Phase A: | 100 mM TEAA in Water |
| Mobile Phase A: | 100 mM TEAA in acetonitrile |
| Gradient: | 10 to 14% B in 10 min |
| Sample: | 10 µL (Agilent Oligonucleotide Resolution Standard (p/n 5190-9028)) |
| LC: | Agilent 1290 Infinity Binary |
Figure 3: An example of an LC/MS analysis of RNA resolution standard containing 14, 17, 20 and 21 mer synthetic oligonucleotides (Courtesy of Agilent, United States)
After reviewing the fundamentals of column chemistry and the mobile phase, let's look at some factors to consider when choosing an ion-pairing agent. Your selection will ultimately affect selectivity and resolution. The most widely utilized ion-pairing agent, particularly for optical-detectors, is TEAA. For a single-stranded oligonucleotide analysis, TEA-HFIP is thought to be the most efficient ion-pairing agent in terms of attaining the best LC-MS sensitivity and resolution since HFIP was introduced as a counter ion. The concentration of both TEA and HFIP is crucial for the success of oligonucleotide separations. The role of the triethylammonium cation (the active ion-pairing agent) is well understood. An increase in TEA concentration improves ion-pairing efficiency and, consequently, the separation selectivity. The side effect of the increased TEA concentration is the rise of mobile phase pH (TEA pKa is 10.7), which may reduce the lifetime of silica-based columns and therefore hybrid columns with a larger usable pH range should be considered. The impact of HFIP on the ion-pairing efficiency of the buffer is less clear. Increasing the HFIP concentration from 100 mM to 400 mM has a positive effect on separation but because HFIP is not an active ion-pairing agent, its effect could only be indirect. The most likely explanation is that the limited solubility of TEA in aqueous HFIP solutions changes the distribution of TEA between the mobile and stationary phases and forces the triethylammonium ion adsorption on the sorbent surface. This, in turn, enhances the ion-pairing retention mechanism and improves the separation performance.
Another important consideration you need to make is whether your mobile phase should be denaturing or not. TEA-HFIP is a denaturing mobile phase, which may or may not be desirable, despite being the gold standard for MS analysis. Fundamentally, we need to attain the appropriate chromatographic resolution, ensure MS compatibility if the analysis calls for it, and decide if the mobile phase needs to be denaturing or non-denaturing. For example, small interfering RNAs (siRNAs) usually occur as oligonucleotide duplexes. They can be analysed under non-denaturing conditions so that duplex formation is assured, and excessive single strands are detected. Alternatively, analysis of siRNAs can be conducted under denaturing conditions so that both sense and antisense strands are monitored.
| Ion Pairing Agent | Buffering Acid | Abbreviation |
| Triethylammonium | Acetate | TEAA |
| Triethylammonium | Bicarbonate | TEAB |
| Di-n-butylammonium | Acetate | DBAA |
| Dimethylbutylammonium | Acetate | DMBAA |
| Tributylammonium | Acetate | TBAA |
| Tripropylammonium | Acetate | TPAA |
| Hexylammonium | Acetate | HAA |
| Triethylammonium | Hexafluoroisopropanol | TEA-HFIP |
Table 2: Partial list of ion-pairing reagents that have been successfully used for synthetic oligonucleotide LC separations.
The type and concentration of ion-pairing buffer have a big impact on the MS intensity. The highest MS sensitivity is usually obtained in TEA-HFIP buffer/methanol systems (acetonitrile cannot be utilized with TEA-HFIP buffer because of its immiscibility). For this reason, even if DBAA and TEAA do not yield the same high resolution and sensitivity as TEA-HFIP, they are nevertheless valuable buffer systems. Ion-pairing reagents with hydrophobic moieties and a positive charge include TEA. Longer retention times and improved resolution are a result of their hydrophobic interaction.


| Column: | Hydrosphere C18 (3 µm, 12 nm) 50 x 2.0 mm ID |
| Part Number: | HS12S03-0502WT |
| Flow rate: | 0.2 mL/min |
| Detection: | UV at 269 nm (red), ESI negative mode (orange) |
| Temperature: | 35°C |
| Injection: | 5 µL (25 pmol/mL) |
| Sample: | Oligodeoxythymidylic acid [d(pT)2-20] |
Figure 4: The effect of salt concentration on the separation and signal intensity of oligonucleotides when comparing TEAA and DBA as alternatives to HFIP-TEA (Courtesy of YMC Europe GmbH, Dinslaken, Germany)
Selecting Mobile phase and Column Temperature
Biomolecules often demonstrate improved peak shape and higher resolution due to increased rates of mass transfer and distribution velocity with rising column temperature. Denaturation of a siRNA duplex produces two peaks of single-stranded RNA, which are visible at 40°C or above. This HPLC technique is called ‘Denaturing HPLC’, which is frequently employed in the field of gene mutation analysis.
Ionic strength (type and concentration), pH, and polarity all have an impact on the denaturation of duplex DNA or RNA. Depending on the properties of the target analyte and the goal of the analysis, analytical conditions should be optimized.
High temperatures and neutral buffers with amino ion-pairing agents work well together in denaturing HPLC, or for high throughput analyses of oligonucleotides. However, because of their reduced stability (silica dissolution at higher pH limit, combined with elevated temperatures), silica-based reversed phase columns are seldom employed under these circumstances; in such cases, hybrid particle columns should be taken into consideration. Hybrid particles contain an organic/inorganic technology, which imparts the high pH dissolution resistance of polymers into the silica particle backbone, without sacrificing any of the silica particles structural integrity or efficiency.


| Column: | YMC Triart C18 (1.9 µm, 12 nm) 100 x 2.0 mm ID |
| Part Number: | TA12SP9-1002PT |
| Flow rate: | 0.2 mL/min |
| Detection: | UV at 269 nm |
| Injection: | 1 µL (5 nmol/mL) |
| System: | Agilent 1290 |
| Condition A Eluent: | A) 10 mM DBAA (pH 6.0) B) methanol |
| Gradient: | 35 – 60% B (0 – 15 min) |
| Condition B Eluent: | A) 20 mM TEAA (pH 7.0) B) acetonitrile |
| Gradient: | 5 – 12 % B (0 – 20 min) |
Figure 5: The separation of siRNA duplex which was compared using different mobile phase conditions at various temperatures (Courtesy of YMC Europe GmbH, Dinslaken, Germany)
Selecting Inert Column Hardware
For many years, stainless steel has been the material of choice for chromatographic columns and systems. These materials are durable, but because they are metals, some analytes that adsorb to those surfaces may have poor peak shapes and low recoveries. Although the corrosion resistance of newer materials like titanium and nickel-cobalt alloys have been used to minimize interactions, analytes can still be absorbed by them. Another material that has been utilised in place of stainless steel is polyether ether ketone (PEEK), however it has disadvantages in terms of pressure (i.e. PEEK is less pressure tolerant and therefore is restricted to larger particle size sorbents and lower pressure instrumentation) and incompatibility with certain mobile phase solvents.
In addition to phosphate groups, as oligonucleotides increase in complexity, they can also include other functional groups that are rich in electrons. These groups can bond to metal surfaces since they have an affinity* for them, necessitating lengthy passivation procedures to get rid of bad peak shape and enhance recovery.
*Metal surfaces (metal oxide surfaces) are positively charged in neutral and acidic pH environments, and ionic interactions with passing molecules can lead to their non-specific binding and adsorption to the surface, especially strong acidic moieties like sulfates, phosphates and stronger carboxylic acids with low pKa and several COOH groups. This happens for stainless steel, titanium and other alloys as well.
New, inert column hardware can eliminate the requirement for extensive and time-consuming passivation, improve recovery and peak shape and ultimately, increase confidence in the data produced.


| Column: | YMC-Accura Triart Bio C4 (3 µm, 30 nm) 100 x 2.1 mm ID |
| Part Number: | TA30S03-10Q1PTC |
| Eluent: | A) 50 mM TEAA (pH 7.0)/acetonitrile (95/5) B) 50 mM TEAA (pH 7.0)/acetonitrile (50/50) |
| Flow rate: | 0.2 mL/min |
| Temperature: | 80°C |
| Detection: | UV at 254 nm |
| Injection: | 1 µL (0.25 mg/mL) |
| Sample: | 100 – 1000 bases (CenturyTM-Plus RNA Markers) |
Figure 6: A comparison between normal and inert column hardware, clearly showing improved performance from the first injection, improved peak shape and sensitivity (Courtesy of YMC Europe GmbH, Dinslaken, Germany)
Ideal choice for challenging analytes such as phosphorothioate oligonucleotides


| Column: | YMC-Accura Triart Bio C18 (1.9 µm, 30 nm) 50 x 2.1 mm ID |
| Part Number: | TA30SP9-05Q1PTC |
| Eluent: | A) 15 mM triethylamine – 400 mM HFIP B) methanol |
| Gradient: | 8 – 18 % B (0 – 10 min) |
| Sample: | All PS RNA 20mer (1) (5’-U^C^A^U^C^A^C^U^G^A^A^U^A^C^C^A^A^U-3’) All PS RNA 21mer (2) (5’G^U^C^A^U^C^A^C^A^C^U^G^A^A^U^A^C^C^A^A^U-3’) ^=Phosphorothioate |
| Detection: | UV at 260 nm |
| Injection: | 1 µL |
| Flow Rate: | 0.42 mL/min |
| Temperature: | 65°C |
Figure 7: Shows the improvement in sensitivity and recovery achieved when using bio-inert hardware (Courtesy of YMC Europe GmbH, Dinslaken, Germany)
Summary and Best Practices
The analysis of oligonucleotides by reversed phase HPLC is a complex and the separation and resolution achieved are the result of numerous factors. The type and concentration of the buffer are crucial elements in IP-RP. Oligonucleotide separations are also greatly influenced by temperature, column hardware, and chemical modifications made to the stationary phase.
Here is a summary of some of the best practices for using IP-RP liquid chromatography to characterise oligonucleotides. Column selection, separation temperature, pH optimization, pore size selection, mobile phase selection, considerations for speedy method development, purification recommendations, and well-liked buffer recipes are all covered by these best practices. Following these best practices helps ensure robust methods, enabling the delivery of consistently high-quality oligonucleotides for therapeutic or diagnostic applications.
- Consider the use of hybrid particle or polar silica particle columns with inert column hardware.
- Perform oligonucleotide separations at elevated pH and temperature for best results.
- Select a suitable pore size for the oligonucleotide separation process. Appropriate pore size selection allows for optimal analyte diffusivity, which optimizes oligonucleotide-bonded phase ligand interaction. Peak shape is enhanced by improved bonded phase ligand interactions.
- Choose an appropriate mobile phase, considering intended use (LC-UV or LC-MS), nature and concentration of the buffer, cost (HFIP can be expensive) and choice of organic modifier (HFIP is immiscible with AcN).
- Method Development:
- Determine the appropriate strength of the gradient or begin with a scouting gradient.
- Shallower gradients improve resolution but increase analysis time; adjust gradient slope to reach desired separation.
- Increase the flow velocity while decreasing the gradient time (constant gradient volume) proportionately to expedite the separation if speed is a priority. - Guidelines for Purification:
- Choose a column dimension that satisfies the requirements for lab-scale isolation.
- The scale of the synthesis reaction mixture is the primary determinant of the column dimension and operating flow rate.
- To optimize component resolution and recovery of the intended product from undesired failure sequences, it is advised to select a suitable column size for oligonucleotide sample loading. For guidelines on bulk loading, consult the manufacturer. - How to make IP-RP Buffers
- Perform all work in a fume hood.
- All mobile phases should be filtered using a 0.45 µm membrane filter that is compatible with solvents and stored in clean, particle-free bottles.
- Recipes for typical IP-RP buffers are readily available from reagent and column manufacturers.
- Select the highest purity buffer reagents to prevent adduct formations.